SPatt: Statistics for Patterns
A simple tutorial SPatt (work in progress)
Toy example with a binary alphabet
Ribosome binding sites in upstream regions of co-regulated genes
Prosite signature in a set of proteins
Using Scilab to perform computations
Let us consider the toy-example pattern
spatt -a "ab" -p "aba" -m 0 -M ab_m0.markov
Processing this command, SPatt produces the Scilab source file pmc.sci which contains the optimal Markov chain embedding of the problem. We can now load this file into Scilab using:
scilab -nw -f pmc.sci
The following commands are then typed into the Scilab interface:
-->x=poly(0,'x'); M=full(P)+x*full(Q)
M =
0.6 0.4 0 0
0 0.4 0.6 0
0.6 0 0 0.4x
0 0.4 0.6 0
-->aux=M^20; res=aux(starts,final)
res =
2 3 4 5 6 7
0.0177338x + 0.0295794x + 0.0256004x + 0.0146999x + 0.0060711x + 0.0018461x + 0.0004048x
8 9
+ 0.0000601x + 0.0000044x
Using graphviz to visualize NFA and DFA
spatt -a "ab" -p "aba" -m 0 --nfa nfa.dot dot nfa.dot -Gbgcolor="transparent" -Tpng > nfa.png
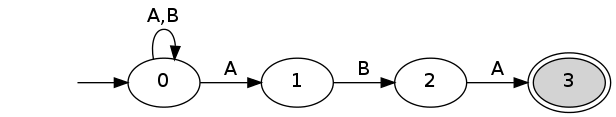
spatt -a "ab" -p "aba" -m 0 --dfa dfa.dot dot dfa.dot -Gbgcolor="transparent" -Tpng > dfa.png
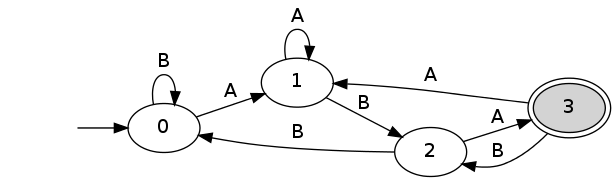
spatt -a "ab" -p "aba" -m 0 -r --dfa dfa_renewal.dot dot dfa.dot -Gbgcolor="transparent" -Tpng > dfa_renewal.png
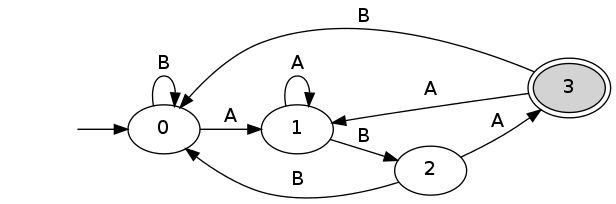
Last edited 01/13/2012